Ready-to-Use Deep Models
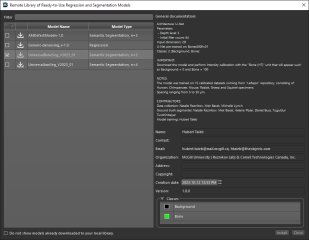
Ready-to-use deep models can greatly accelerate workflows for image enhancement and segmentation tasks. You can browse for and download selected models to your local library from the Remote Library of Ready-to-Use Regression and Segmentation Models dialog, shown below. Downloaded models can be used immediately or fine-tuned by performing additional training with your own datasets.
Remote Library of Ready-to-Use Regression and Segmentation Models dialog
Models are listed as Available for Download ![]() and Installed in Local Library
and Installed in Local Library ![]() in the dialog. In addition, the general documentation provides information about each model’s architecture, parameters, input dimensions, the number of classes for semantic segmentation models, as well as usage notes such as calibration requirements. Models available in the Remote Library of Ready-to-Use Regression and Segmentation Models are listed below.
in the dialog. In addition, the general documentation provides information about each model’s architecture, parameters, input dimensions, the number of classes for semantic segmentation models, as well as usage notes such as calibration requirements. Models available in the Remote Library of Ready-to-Use Regression and Segmentation Models are listed below.
| Model | Description |
|---|---|
| AMDefectModel |
Trained for segmenting pores, material, and background on calibrated datasets of additively manufactured metal cubes.
Architecture: U-Net
Note Input data must be calibrated such that the background = 0 and material = 100. Reference Taute, C., Moller, H., Du Plessis, A., Tshibalanganda, M. and Leary, M., 2021. Characterization of additively manufactured AlSilOMg cubes with different porosities. Journal of the Southern African Institute of Mining and Metallurgy, 121(4), pp.143-150. |
| Generic-denoising |
This model was trained for general denoising with the following settings:
Architecture: U-Net
|
| SEM-Denoise |
This model was trained for denoising SEM datasets with the following settings:
Architecture: U-Net
|
| UniversalBoneSeg |
This model can be used for segmenting bone and was pre-trained on Bones500Kv01 and then fine-tuned on 15 calibrated datasets consisting of human, chimpanzee, mouse, rabbit, sheep, and squirrel specimens. Spacing values ranged from 5 to 50 micrometers. The settings of the model are as follows:
Architecture: U-Net
Note Input data must be calibrated such that background = 0 and bone = 100. |
| UniversalJawSeg |
This model can be used for segmenting bone, dentin, and enamel and was pre-trained on ORS500Kv01 and then fine-tuned on four calibrated datasets from the repository "LeRepo". Spacing values ranged from 15 to 50 micrometers. The settings of the model are as follows:
Architecture: U-Net
Note Input data must be calibrated such that background = 0 and enamel = 100. |
-
Do one of the following:
-
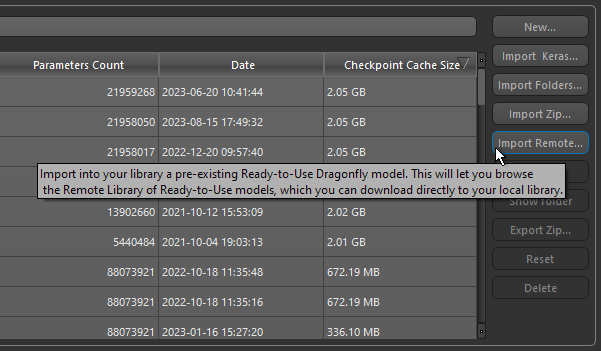
Open the Deep Learning Tool and then click the Import Remote button on the Model Overview panel.
-
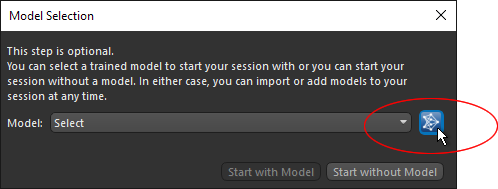
Launch the Segmentation Wizard and then click the Open Remote Library button on the Model Selection dialog, as shown below.
Note You can also open the Remote Library of Ready-to-Use Segmentation Models dialog by clicking the Open Remote Library button on the Models tab, as shown below.
-
Open the Segment with AI panel and then click the Open Remote Library button, as shown below.
-
Open the Filter with AI panel and then click the Open Remote Library button, as shown below.
-
-
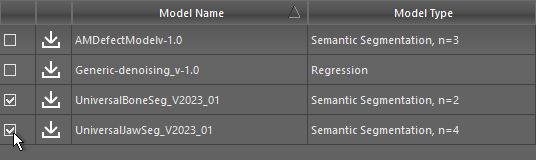
Check the model(s) that you want to download in the Remote Library of Ready-to-Use Models dialog, as shown below.
-
Click the Install button.
The selected model(s) are downloaded to your local library for immediate use or for fine-tuning with your own training data.